There’s a worldwide scarcity of entry to medical imaging skilled interpretation throughout specialties together with radiology, dermatology and pathology. Machine studying (ML) know-how will help ease this burden by powering instruments that allow docs to interpret these photos extra precisely and effectively. Nevertheless, the event and implementation of such ML instruments are sometimes restricted by the provision of high-quality knowledge, ML experience, and computational sources.
One option to catalyze the usage of ML for medical imaging is by way of domain-specific fashions that make the most of deep studying (DL) to seize the knowledge in medical photos as compressed numerical vectors (known as embeddings). These embeddings characterize a kind of pre-learned understanding of the necessary options in a picture. Figuring out patterns within the embeddings reduces the quantity of knowledge, experience, and compute wanted to coach performant fashions as in comparison with working with high-dimensional data, reminiscent of photos, straight. Certainly, these embeddings can be utilized to carry out quite a lot of downstream duties inside the specialised area (see animated graphic under). This framework of leveraging pre-learned understanding to unravel associated duties is just like that of a seasoned guitar participant shortly studying a brand new tune by ear. As a result of the guitar participant has already constructed up a basis of talent and understanding, they’ll shortly choose up the patterns and groove of a brand new tune.
As a way to make the sort of embedding mannequin obtainable and drive additional growth of ML instruments in medical imaging, we’re excited to launch two domain-specific instruments for analysis use: Derm Foundation and Path Foundation. This follows on the robust response we’ve already obtained from researchers utilizing the CXR Foundation embedding instrument for chest radiographs and represents a portion of our increasing analysis choices throughout a number of medical-specialized modalities. These embedding instruments take a picture as enter and produce a numerical vector (the embedding) that’s specialised to the domains of dermatology and digital pathology photos, respectively. By operating a dataset of chest X-ray, dermatology, or pathology photos by way of the respective embedding instrument, researchers can get hold of embeddings for their very own photos, and use these embeddings to shortly develop new fashions for his or her functions.
Path Basis
In “Domain-specific optimization and diverse evaluation of self-supervised models for histopathology”, we confirmed that self-supervised studying (SSL) fashions for pathology photos outperform conventional pre-training approaches and allow environment friendly coaching of classifiers for downstream duties. This effort targeted on hematoxylin and eosin (H&E) stained slides, the principal tissue stain in diagnostic pathology that allows pathologists to visualise mobile options beneath a microscope. The efficiency of linear classifiers skilled utilizing the output of the SSL fashions matched that of prior DL fashions skilled on orders of magnitude extra labeled knowledge.
Resulting from substantial variations between digital pathology photos and “pure picture” photographs, this work concerned a number of pathology-specific optimizations throughout mannequin coaching. One key ingredient is that whole-slide images (WSIs) in pathology might be 100,000 pixels throughout (hundreds of occasions bigger than typical smartphone photographs) and are analyzed by consultants at a number of magnifications (zoom ranges). As such, the WSIs are sometimes damaged down into smaller tiles or patches for laptop imaginative and prescient and DL functions. The ensuing photos are info dense with cells or tissue constructions distributed all through the body as an alternative of getting distinct semantic objects or foreground vs. background variations, thus creating distinctive challenges for strong SSL and have extraction. Moreover, bodily (e.g., cutting) and chemical (e.g., fixing and staining) processes used to organize the samples can affect picture look dramatically.
Taking these necessary elements into consideration, pathology-specific SSL optimizations included serving to the mannequin be taught stain-agnostic features, generalizing the mannequin to patches from a number of magnifications, augmenting the information to imitate scanning and picture put up processing, and customized knowledge balancing to enhance enter heterogeneity for SSL coaching. These approaches have been extensively evaluated utilizing a broad set of benchmark duties involving 17 totally different tissue sorts over 12 totally different duties.
Using the imaginative and prescient transformer (ViT-S/16) structure, Path Basis was chosen as one of the best performing mannequin from the optimization and analysis course of described above (and illustrated within the determine under). This mannequin thus supplies an necessary steadiness between efficiency and mannequin dimension to allow invaluable and scalable use in producing embeddings over the various particular person picture patches of huge pathology WSIs.
 |
| SSL coaching with pathology-specific optimizations for Path Basis. |
The worth of domain-specific picture representations may also be seen within the determine under, which reveals the linear probing efficiency enchancment of Path Basis (as measured by AUROC) in comparison with conventional pre-training on pure photos (ImageNet-21k). This contains analysis for duties reminiscent of metastatic breast cancer detection in lymph nodes, prostate cancer grading, and breast cancer grading, amongst others.
 |
| Path Basis embeddings considerably outperform conventional ImageNet embeddings as evaluated by linear probing throughout a number of analysis duties in histopathology. |
Derm Basis
Derm Foundation is an embedding instrument derived from our analysis in making use of DL to interpret images of dermatology conditions and contains our current work that provides improvements to generalize better to new datasets. Resulting from its dermatology-specific pre-training it has a latent understanding of options current in photos of pores and skin situations and can be utilized to shortly develop fashions to categorise pores and skin situations. The mannequin underlying the API is a BiT ResNet-101×3 skilled in two phases. The primary pre-training stage makes use of contrastive studying, just like ConVIRT, to coach on numerous image-text pairs from the internet. Within the second stage, the picture element of this pre-trained mannequin is then fine-tuned for situation classification utilizing medical datasets, reminiscent of these from teledermatology providers.
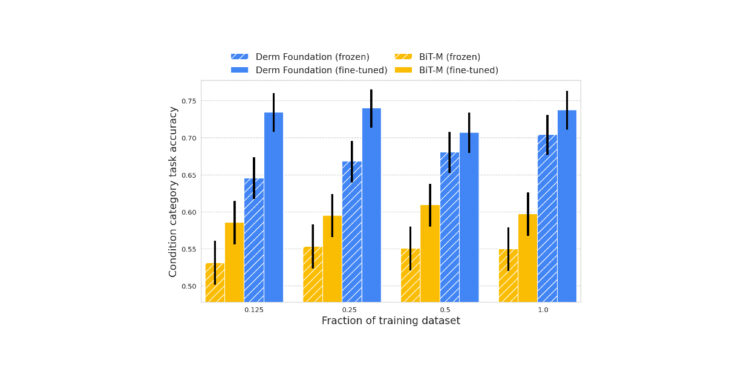
Not like histopathology photos, dermatology photos extra carefully resemble the real-world photos used to coach a lot of immediately’s laptop imaginative and prescient fashions. Nevertheless, for specialised dermatology duties, making a high-quality mannequin should still require a big dataset. With Derm Basis, researchers can use their very own smaller dataset to retrieve domain-specific embeddings, and use these to construct smaller fashions (e.g., linear classifiers or different small non-linear fashions) that allow them to validate their analysis or product concepts. To guage this method, we skilled fashions on a downstream process utilizing teledermatology knowledge. Mannequin coaching concerned various dataset sizes (12.5%, 25%, 50%, 100%) to check embedding-based linear classifiers towards fine-tuning.
The modeling variants thought-about have been:
- A linear classifier on frozen embeddings from BiT-M (a normal pre-trained picture mannequin)
- High quality-tuned model of BiT-M with an additional dense layer for the downstream process
- A linear classifier on frozen embeddings from the Derm Basis API
- High quality-tuned model of the mannequin underlying the Derm Basis API with an additional layer for the downstream process
We discovered that fashions constructed on prime of the Derm Basis embeddings for dermatology-related duties achieved considerably increased high quality than these constructed solely on embeddings or advantageous tuned from BiT-M. This benefit was discovered to be most pronounced for smaller coaching dataset sizes.
Nevertheless, there are limitations with this evaluation. We’re nonetheless exploring how nicely these embeddings generalize throughout process sorts, affected person populations, and picture settings. Downstream fashions constructed utilizing Derm Basis nonetheless require cautious analysis to know their anticipated efficiency within the meant setting.
Entry Path and Derm Basis
We envision that the Derm Basis and Path Basis embedding instruments will allow a variety of use instances, together with environment friendly growth of fashions for diagnostic duties, high quality assurance and pre-analytical workflow enhancements, picture indexing and curation, and biomarker discovery and validation. We’re releasing each instruments to the analysis group to allow them to discover the utility of the embeddings for their very own dermatology and pathology knowledge.
To get entry, please signal as much as every instrument’s phrases of service utilizing the next Google Kinds.
After getting access to every instrument, you should use the API to retrieve embeddings from dermatology photos or digital pathology photos saved in Google Cloud. Accepted customers who’re simply curious to see the mannequin and embeddings in motion can use the supplied instance Colab notebooks to coach fashions utilizing public knowledge for classifying six common skin conditions or figuring out tumors in histopathology patches. We look ahead to seeing the vary of use-cases these instruments can unlock.
Acknowledgements
We want to thank the various collaborators who helped make this work potential together with Yun Liu, Can Kirmizi, Fereshteh Mahvar, Bram Sterling, Arman Tajback, Kenneth Philbrik, Arnav Agharwal, Aurora Cheung, Andrew Sellergren, Boris Babenko, Basil Mustafa, Jan Freyberg, Terry Spitz, Yuan Liu, Pinal Bavishi, Ayush Jain, Amit Talreja, Rajeev Rikhye, Abbi Ward, Jeremy Lai, Faruk Ahmed, Supriya Vijay,Tiam Jaroensri, Jessica Bathroom, Saurabh Vyawahare, Saloni Agarwal, Ellery Wulczyn, Jonathan Krause, Fayaz Jamil, Tom Small, Annisah Um’rani, Lauren Winer, Sami Lachgar, Yossi Matias, Greg Corrado, and Dale Webster.