Transform your machine learning performance by focusing on what matters most
Have you ever wondered why some training samples seem more important than others for your model’s performance? What if I told you there’s a way to automatically identify and prioritize the most valuable training examples based on their similarity to your test data?
Enter SemiDeep a game-changing Python package that implements distance-based sample weighting to supercharge your deep learning models.
Traditional deep learning treats all training samples equally. But in reality:
- Some samples are more representative of your test distribution
- Class imbalance can skew your model’s focus
- Noisy labels can mislead the learning process
- Domain shift between training and test data reduces performance
SemiDeep solves this by giving your model a smarter way to learn.
SemiDeep implements a research-backed approach from the paper “Enhancing Classification with Semi-Supervised Deep Learning Using Distance-Based Sample Weights” . Here’s the core insight:
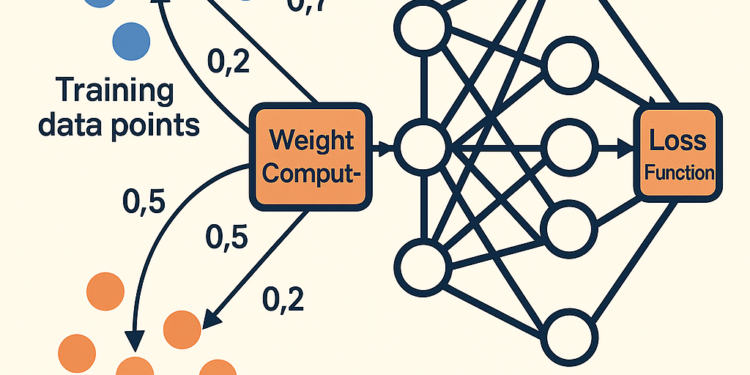
Training samples that are more similar to test samples should have higher influence on the learning process.
The magic happens through this elegant formula:
w_i = (1/M) * Σ_j exp(-λ · d(x_i, x_j'))
Where:
w_iis the weight for training sample id(x_i, x_j')is the distance between training and test samplesλcontrols how quickly influence decays with distance
Let’s dive right into code. First, install the package:
pip install semideep
Here’s a complete example using the breast cancer dataset:
import torch
import torch.nn as nn
from semideep import WeightedTrainer
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
# Load and prepare data
data = load_breast_cancer()
X, y = data.data, data.targetX_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)# Define your model
class SimpleModel(nn.Module):
def __init__(self, input_dim):
super().__init__()
self.layers = nn.Sequential(
nn.Linear(input_dim, 64),
nn.ReLU(),
nn.Dropout(0.2),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 2)
)def forward(self, x):
# Here's where the magic happens
return self.layers(x)
model = SimpleModel(input_dim=X_train.shape[1])
trainer = WeightedTrainer(
model=model,
X_train=X_train,
y_train=y_train,
X_test=X_test,
weights="distance", # Enable distance-based weighting
distance_metric="cosine",
lambda_=0.8,
epochs=100,
learning_rate=0.001,
batch_size=32
)# Train and evaluate
history = trainer.train()
metrics = trainer.evaluate(X_test, y_test)
print(f"Test accuracy: {metrics['accuracy']:.4f}")
That’s it! With just a few lines of code, you’ve implemented sophisticated sample weighting.
Not sure which distance metric works best for your data? SemiDeep has you covered:
from semideep import auto_select_distance_metric
# Let SemiDeep choose based on your data characteristics
best_metric = auto_select_distance_metric(X_train)
print(f"Recommended metric: {best_metric}")
from semideep import select_best_distance_metric
def create_model():
return SimpleModel(input_dim=X_train.shape[1])best_metric, best_lambda, best_score = select_best_distance_metric(
model=create_model(),
X_train=X_train,
y_train=y_train,
X_test=X_test,
metrics=['euclidean', 'cosine', 'hamming', 'jaccard'],
lambda_values=[0.5, 0.7, 0.8, 0.9, 1.0],
verbose=True
)print(f"Best combination: {best_metric} with λ={best_lambda}")
For maximum flexibility, compute weights manually and integrate them into your custom training loop:
from semideep import WeightComputer, WeightedLoss
# Compute weights separately
weight_computer = WeightComputer(
distance_metric="euclidean",
lambda_=0.8
)
weights = weight_computer.compute_weights(X_train, X_test)# Create weighted loss function
criterion = WeightedLoss(nn.CrossEntropyLoss())# Your custom training loop
model = SimpleModel(input_dim=X_train.shape[1])
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)X_train_tensor = torch.FloatTensor(X_train)
y_train_tensor = torch.LongTensor(y_train)
weights_tensor = torch.FloatTensor(weights)for epoch in range(100):
model.train()
optimizer.zero_grad()
outputs = model(X_train_tensor)
loss = criterion(outputs, y_train_tensor, weights_tensor)
loss.backward()
optimizer.step()
Let’s see SemiDeep in action with a challenging imbalanced dataset:
from sklearn.datasets import make_classification
from collections import Counter
# Create heavily imbalanced data (10:1 ratio)
X, y = make_classification(
n_samples=1000, n_features=20, n_informative=15,
n_redundant=5, n_classes=2, weights=[0.1, 0.9],
random_state=42
)X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42, stratify=y
)print(f"Training class distribution: {Counter(y_train)}")
# Output: Counter({1: 630, 0: 70}) - Highly imbalanced!# Train with SemiDeep
model = SimpleModel(input_dim=X_train.shape[1])
trainer = WeightedTrainer(
model=model,
X_train=X_train,
y_train=y_train,
X_test=X_test,
weights="distance",
distance_metric="cosine",
lambda_=0.8,
epochs=100
)trainer.train()
metrics = trainer.evaluate(X_test, y_test)
Always compare your SemiDeep results against a baseline:
# Baseline model (no weighting)
baseline_model = SimpleModel(input_dim=X_train.shape[1])
baseline_trainer = WeightedTrainer(
model=baseline_model,
X_train=X_train,
y_train=y_train,
X_test=X_test,
weights=None, # No weighting
epochs=100
)
baseline_trainer.train()
baseline_metrics = baseline_trainer.evaluate(X_test, y_test)
# SemiDeep model
semideep_model = SimpleModel(input_dim=X_train.shape[1])
semideep_trainer = WeightedTrainer(
model=semideep_model,
X_train=X_train,
y_train=y_train,
X_test=X_test,
weights="distance",
distance_metric="cosine",
lambda_=0.8,
epochs=100
)
semideep_trainer.train()
semideep_metrics = semideep_trainer.evaluate(X_test, y_test)# Calculate improvements
for metric in semideep_metrics:
if metric != 'val_loss':
improvement = semideep_metrics[metric] - baseline_metrics[metric]
percent = improvement / max(baseline_metrics[metric], 1e-10) * 100
print(f"{metric}: +{improvement:.4f} ({percent:+.2f}%)")
SemiDeep shines in these scenarios:
- Limited labeled data: Make the most of every training sample
- Class imbalance: Automatically focus on underrepresented classes
- Noisy labels: Reduce the impact of mislabeled examples
- Domain shift: Bridge the gap between training and test distributions
- Transfer learning: Adapt pre-trained models to new domains
- Simple Integration: Add distance-based weighting with just one parameter
- Automatic Optimization: Let SemiDeep find the best distance metric and parameters
- Flexible Usage: From plug-and-play to full customization
- Research-Backed: Based on peer-reviewed methodology
- Real Performance Gains: Measurable improvements across various scenarios
Ready to boost your models? Install SemiDeep and give it a try:
pip install semideep
Check out the GitHub repository for more examples and documentation.